High-grade serous tubo-ovarian cancer refined with single-cell RNA-sequencing: specific cell subtypes influence survival and determine molecular subtype classification
High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: specific cell subtypes influence survival and determine molecular subtype classification
-
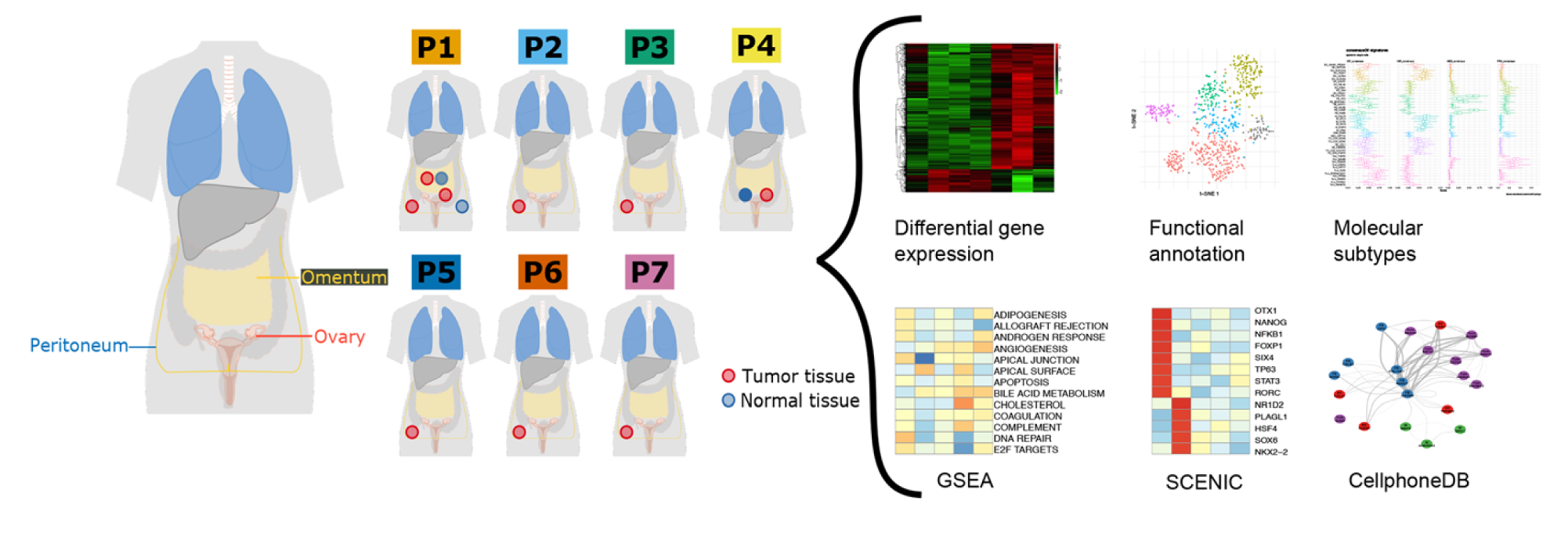
High-grade serous tubo-ovarian cancer (HGSTOC) is characterised by extensive inter- and intratumour heterogeneity, resulting in persistent therapeutic resistance and poor disease outcome. Molecular subtype classification based on bulk RNA sequencing facilitates a more accurate characterisation of this heterogeneity, but the lack of strong prognostic or predictive correlations with these subtypes currently hinders their clinical implementation. Stromal admixture profoundly affects the prognostic impact of the molecular subtypes, but the contribution of stromal cells to each subtype has poorly been characterised. Increasing the transcriptomic resolution of the molecular subtypes based on single-cell RNA sequencing (scRNA-seq) may provide insights in the prognostic and predictive relevance of these subtypes. We performed scRNA-seq of 18,403 cells unbiasedly collected from 7 treatment-naive HGSTOC tumours. For each phenotypic cluster of tumour or stromal cells, we identified specific transcriptomic markers. We explored which phenotypic clusters correlated with overall survival based on expression of these transcriptomic markers in microarray data of 1467 tumours. By evaluating molecular subtype signatures in single cells, we assessed to what extent a phenotypic cluster of tumour or stromal cells contributes to each molecular subtype. We identified 11 cancer and 32 stromal cell phenotypes in HGSTOC tumours. Of these, the relative frequency of myofibroblasts, TGF-β-driven cancer-associated fibroblasts, mesothelial cells and lymphatic endothelial cells predicted poor outcome, while plasma cells correlated with more favourable outcome. Moreover, we identified a clear cell-like transcriptomic signature in cancer cells, which correlated with worse overall survival in HGSTOC patients. Stromal cell phenotypes differed substantially between molecular subtypes. For instance, the mesenchymal, immunoreactive and differentiated signatures were characterised by specific fibroblast, immune cell and myofibroblast/mesothelial cell phenotypes, respectively. Cell phenotypes correlating with poor outcome were enriched in molecular subtypes associated with poor outcome. In conclusion, we used scRNA-seq to identify stromal cell phenotypes predicting overall survival in HGSTOC patients. These stromal features explain the association of the molecular subtypes with outcome but also the latter's weakness of clinical implementation. Stratifying patients based on marker genes specific for these phenotypes represents a promising approach to predict prognosis or response to therapy.
-
High-grade serous tubo-ovarian cancer refined with single-cell RNA-sequencing: specific cell subtypes influence survival and determine molecular subtype classification., Olbrecht et al., Genome Medicine 2021.
Data availability
Raw sequencing reads of the scRNA-seq experiments have been deposited in the European Genome-phenome Archive under accession number EGAS00001004987 (EGA; https://ega-archive.org/studies/EGAS00001004987). Requests for accessing raw sequencing reads will have to be reviewed by the UZLeuven-VIB data access committee. Any data shared will only be released prior to a Data Transfer Agreement that will have to include the necessary conditions to guarantee protection of personal data (according to European GDPR law).
Alternatively, a download of the read count data, meta-data and Seurat scripts is more readily available from this website.
The count matrix of the pan-cancer blueprint data from Qian et al. is available here.
