Data Access
CD4+ T cell activation distinguishes response to anti-PD-L1+anti-CTLA4 therapy from anti-PD-L1 monotherapy
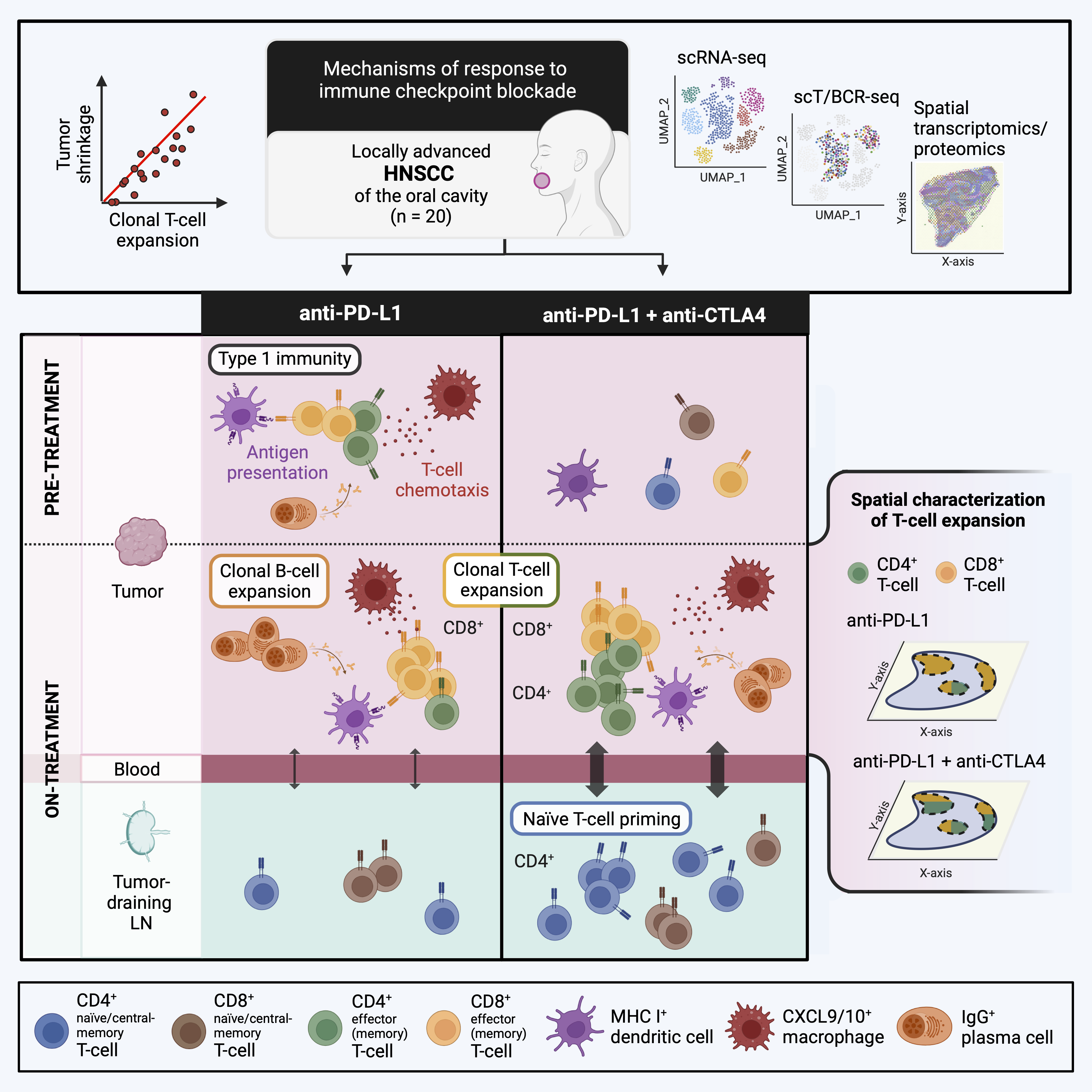
Cancer patients often receive a combination of PD-(L)1 and CTLA4 inhibitors, but the specific contribution of adding anti-CTLA4 to anti-PD-L1 alone is unclear. We conducted a window-of-opportunity study in head and neck squamous cell carcinoma (HNSCC) involving anti-PD-L1 monotherapy versus anti-PD-L1 plus anti-CTLA4 combination. Single-cell profiling of on- versus pre-treatment biopsies identified T cell expansion as early response marker. In tumors, anti-PD-L1 triggered expansion of mostly CD8+ T cells while combination therapy facilitated expansion of both CD4+ and CD8+ T cells. Particularly, CD4+ naïve/central memory T cells (Tn/cm cells) expanded towards activated T helper 1 (Th1) cells. Spatial analysis of expanding T cells confirmed CD4+ and CD8+ expanding T cells to be co-localized and surrounded by dendritic cells expressing T cell homing factors or antibody-producing plasma cells. T cell receptor tracing suggested anti-CTLA4 but not anti-PD-L1 to trigger trafficking of CD4+ Tn/cm cells from tumor-draining lymph nodes, via blood, towards activated Th1 cells in tumors. We reveal a critical role for CD4+ T cell activation and CD4+ T cell recruitment from tdLNs during early response to anti-PD-L1 plus anti-CTLA4.
CD4+ T cell activation distinguishes response to anti-PD-L1+anti-CTLA4 therapy from anti-PD-L1 monotherapy in head and neck squamous cell carcinoma. Franken et al. – 2024 – Immunity
Data and Code Availability
- A download of the read count data per individual patient is publicly available from this website (see below). Raw sequencing reads of the scRNA-seq, scTCR-seq, and scBCR-seq experiments were deposited under restricted access in the European Genome-phenome Archive (EGA; study submission number: EGA50000000033). Requests for accessing raw sequencing reads will be reviewed by the UZ Leuven-VIB data access committee. Any data shared will be released via a Data Transfer Agreement that will include the necessary conditions to guarantee protection of personal data (according to European GDPR law). Raw sequencing reads of the spatial transcriptomics data were deposited in the KU Leuven Research Data Repository (doi:10.48804/992X8C). Please note that sample and patient identifiers have been anonymized and shuffled in these data releases and may not match those described in the paper or the supplement.
- All original code has been deposited at GitHub and is publicly available as of the date of publication.
- Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
