Expertise
Expertise
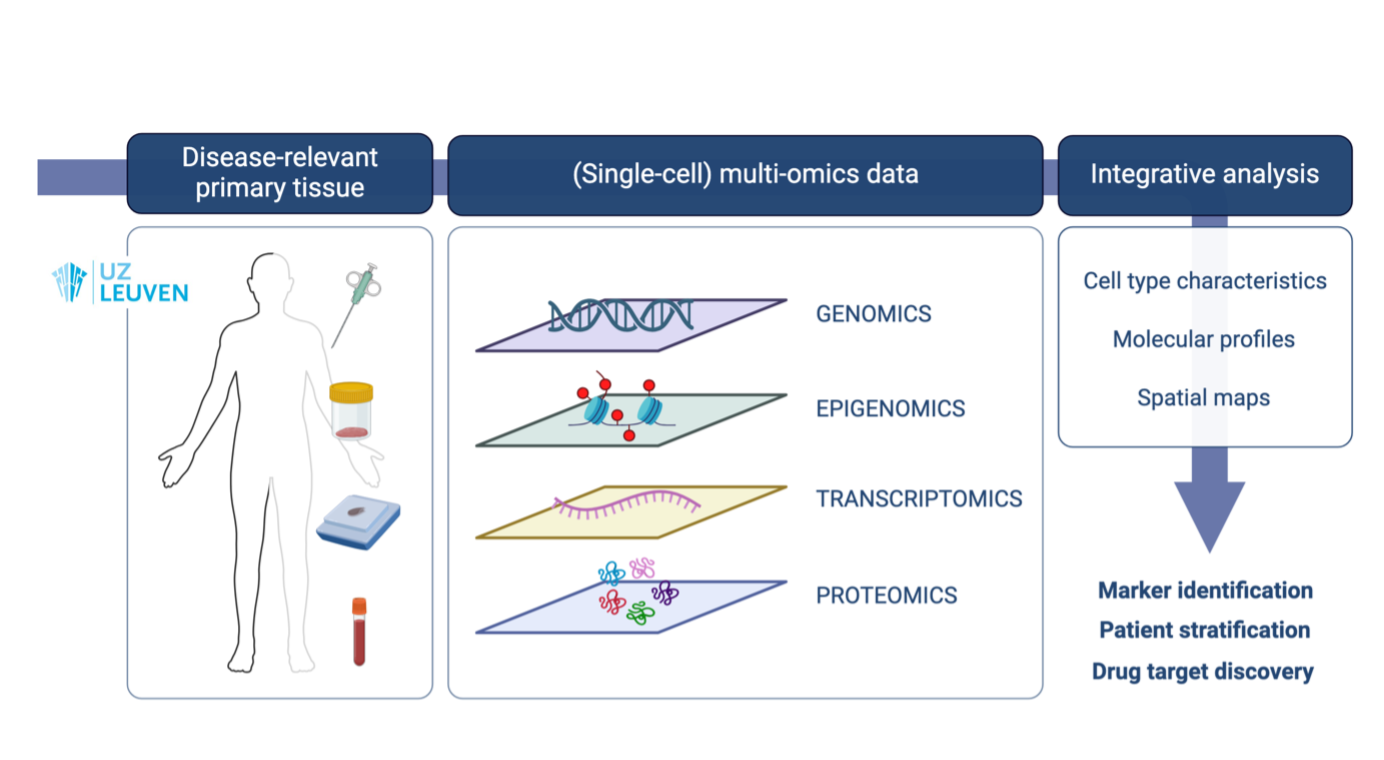
Access to high-throughput next-generation technologies
Our laboratory boasts unparalleled proficiency in leveraging high-throughput (single-cell) (epi)genomic, transcriptomic, and proteomic profiling methodologies. We excel in conducting state-of-the-art multi-omics investigations, which are often fueled by the swift evolution of pioneering yet costly technologies.
Within our facility, we offer direct and extensive utilization of diverse sequencing platforms, encompassing the Illumina iSCAN, MiSeq, NextSeq, HiSeq2500, HiSeq4000, and NovaSeq6000. This comprehensive suite empowers us to swiftly execute whole-genome sequencing (WGS), targeted (re-)sequencing (such as exome sequencing), (targeted) bisulphite sequencing [5(h)mC-Seq], and RNA sequencing. Our proficiency extends to accommodating various sample types, including fresh frozen (tumor) tissue, FFPE-based (tumor) tissue, bronchoalveolar lavage fluid, and liquid biopsies.
Furthermore, through our involvement in the single-cell accelerator initiative at VIB, our laboratory enjoys early access to the latest single-cell multi-omics sequencing technologies, encompassing scRNA-seq, snRNA-seq, ATAC-seq, DNA-seq, CITE-seq, and CROP-seq methodologies. Additionally, we oversee the management of the 10X Genomics technology and Illumina NovaSeq system for all single-cell RNA sequencing instruments at UZ Leuven and KU Leuven via the Genomics Core (https://www.genomicscore.be/).
To enable the spatial characterization of tumors, we have made strategic investments in spatial technologies, including untargeted spatial transcriptomics platforms (Visium (10x Genomics), Xenium (10x Genomics), and MERSCOPE (Vizgen)), as well as a targeted spatial proteomics platform (PhenoCycler-Fusion 2.0 (Akoya Biosciences)). Additionally, we are pioneering the implementation of combined spatial RNA and TCR profiling at subcellular resolution through the Nova-ST workflow, adapted by Poovathingal et al. These cutting-edge innovations empower us to unravel the complex spatial architecture of biological specimens with unparalleled precision and depth, unlocking novel perspectives in cellular and molecular biology.
Advanced multi-omics data analysis by cutting-edge bioinformatics and data integration strategies
Within our laboratory, we possess the expertise needed to harness cutting-edge bioinformatics and data integration techniques for multi-omics analysis. Our team is proficient in clustering omics-type data into molecular subtypes and developing tailored classifiers suitable for clinical applications.
A significant investment has been made to attract top-tier computational biologists, who spearhead the analysis and integration of extensive genomic datasets. Currently, our team comprises 3 experienced postdocs, > 10 PhD students (including medical doctors), and 6 dedicated staff scientists, all focusing on dissecting numerous large-scale multi-omics datasets.
To facilitate these extensive computational analyses, our laboratory boasts a state-of-the-art computing infrastructure, equipped with over 340 processors and 66 terabytes of storage capacity. Additionally, we have seamless access to the Flemish Supercomputer Center (Vlaams Supercomputer Centrum, VSC), ensuring access to high-performance computing resources when needed. This robust technological foundation enables us to navigate complex omics data landscapes with precision and efficiency, empowering us to extract invaluable insights crucial for advancing clinical understanding and personalized medicine approaches.
Our technologies and bioinformatics pipelines are accessible collaboratively (contact: Diether Lambrechts) or on a fee-for-service basis (https://www.genomicscore.be/).
